Postdoc Projects (2013-2015)
Fatty Acid Source Tracking Algorithm in R (FASTAR)
A fatty acid-based Bayesian mixing model approach for quantitative estimation basal resource utilization by herbivorous invertebrates. I have worked on several applications of the approach and am currently contributing to refinement of the initial published methods in new collaborations.

Under-Ice Ecology of Lakes
There is growing interest in the ecology of lakes during periods of ice cover, but our general understanding of under-ice productivity and biology in lakes is still limited. For my postdoc work with Dr. Stephanie Hampton, we organized a large collaborative effort to synthesize global under-ice biological data.
Food Quality of Algal Resources
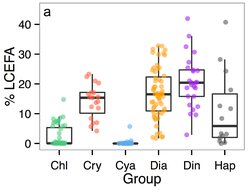
For my postdoc with Dr. Monika Winder, we summarized the long chain essential fatty acid content (LCEFA) from six algal groups, using hundreds of published phytoplankton fatty acid profiles. We applied average group LCEFA content to a multi-decadal time phytoplankton time series to model fatty acid dynamics in a freshwater ecosystem. The work (and the data) is published at PLoS ONE.
Modeling Zooplankton Diet with Biomarkers
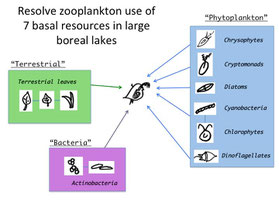
For my postdcoc with Dr. Paula Kankaala in Finland, I used a novel fatty acid mixing model, paired with an extensive series of feeding trials with Daphnia, to estimate the contribution of diverse phytoplankton, bacteria, and terrestrial-originated detritus to cladocera in 5 large boreal lakes (Freshwater Bio. 2014).

What Resources Support Intertidal Isopods?
Isopods are generalist herbivores that can regulate algal communities and transfer energy from primary producers to upper trophic levels. However, direct observation, stomach contents, and existing biomarker methods cannot effectively track what isopods eat. We quantified isopod algal resource use (MEPS 2014) with a fatty-acid mixing model based on feeding trial data. We found that individual isopod diets can vary markedly from within sites, but also that isopods are generally supported by the locally abundant algae.
 Aaron W. E. Galloway
Aaron W. E. Galloway